In the current biotechnology and medicine era, genomic data analysis is the compass guiding researchers through our genetic code.
This comprehensive guide aims to explain genomic data analysis along with Python and R coding, breaking down each step into digestible portions.
Before diving into the nitty-gritty of genomic data analysis, let’s grasp the concept of genomic data, its formats and significance of genomic data analysis.
Explore more about role of 20 Essential Python Codes for Bioinformatics Beginners.
What is Genomic Data?
Genomic data is the code embedded in DNA sequences of living organisms, serving as the fundamental blueprint of life. It contains the entire set of genetic instructions that govern the development, functioning, and characteristics of an organism. This data includes the sequence of nucleotide bases – adenine (A), thymine (T), cytosine (C), and guanine (G) – arranged in a specific order, forming genes that act as the building blocks of our biological existence.
Genomic data is the molecular language that scientists decode to understand the inherited traits, the origins of diseases, and the evolutionary paths of diverse species

What are Common Genomic Data Formats?
Common genomic data formats include FASTQ, which holds raw sequence data, and BAM, a binary version of the Sequence Alignment/Map (SAM) format used for storing aligned sequence reads. Variant Call Format (VCF) is another vital format, specifically designed to represent genetic variations detected during genomic data analysis.
What is Genomic Data Analysis and Why it is Important?
Genomic data analysis is the systematic examination of the vast amount of genetic information contained within an organism’s DNA. It involves employing computational techniques and specialized tools to explore the genomic code.
The primary goal of genomic data analysis is to extract meaningful insights from the genetic data, understanding the functions of genes, identifying variations, and exploring the relationships between different elements within the genome. This process serves as a crucial bridge between the raw genetic information and actionable knowledge. It allows scientists, researchers, and healthcare professionals to comprehend the genetic basis of various phenomena, such as inherited traits, diseases, and evolutionary patterns. In essence, genomic data analysis provide a deeper understanding of life’s molecular intricacies and pave the way for advancements in medicine, biology, and genetics.
What Steps are Involved in Genomic Data Analysis?
Genomic data analysis involves a number of steps which are explained here in detail with python and R coding examples
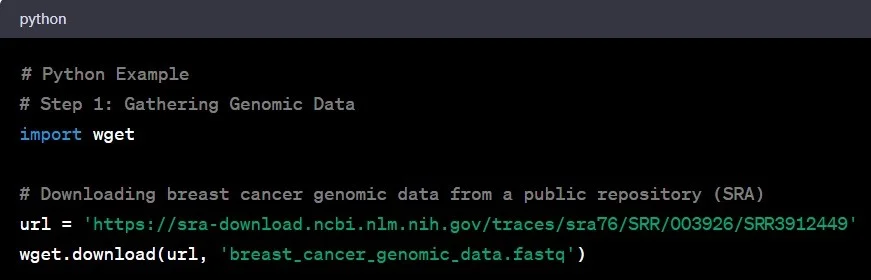
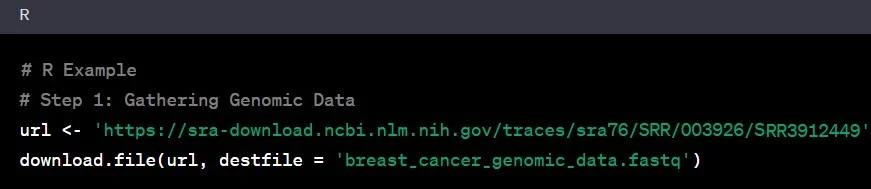
Step 1: Gathering Genomic Data
Think of genomic data as the raw material for our genetic detective work. This data comes in various formats – FASTQ, BAM, or VCF. Imagine stepping into a vast library of genetic information, akin to NCBI’s Sequence Read Archive (SRA), where scientists worldwide deposit their genomic findings. Let’s say you’re interested in studying genetic variations in breast cancer. You’d download the relevant genomic data from SRA or collaborate with a renowned research institution specializing in cancer genomics.
Example: You access the SRA database and download genomic data from a study on breast cancer patients.
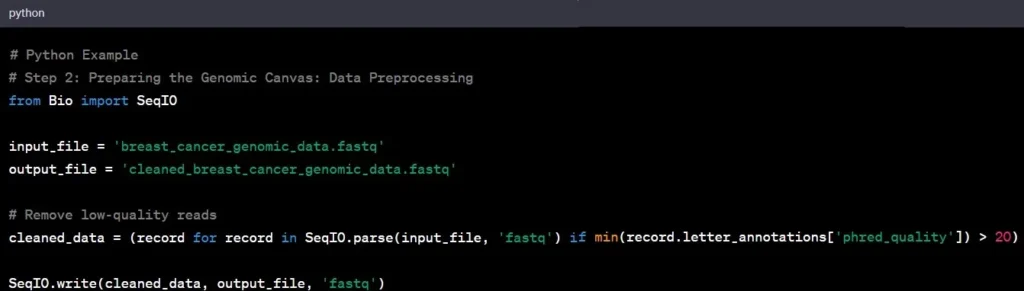
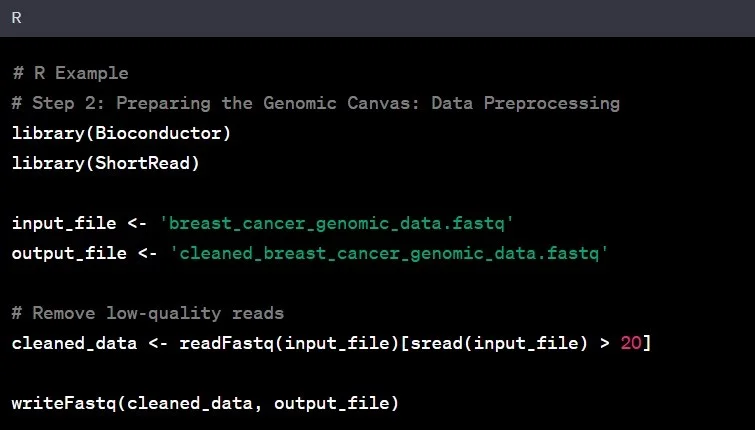
Step 2: Data Preprocessing
Now that we have our raw data, it’s time to clean it up – think of it as preparing a canvas before painting. This step, known as data preprocessing, involves removing noise, correcting errors, and ensuring the data’s overall quality.
Example: In our breast cancer study, you would eliminate low-quality reads, ensuring that the remaining data is reliable for subsequent analysis.
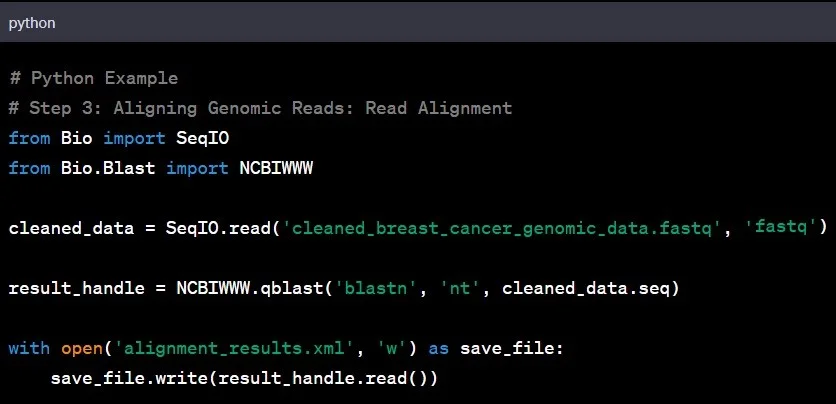
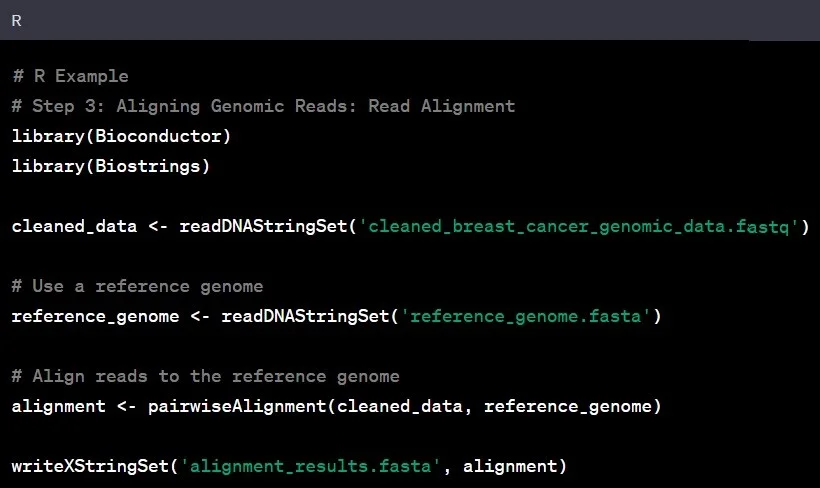
Step 3: Read Alignment
Reading the genomic sequence is the next step in genomic data analysis. Aligning the cleaned genomic data to a reference genome helps identify variations and understand the larger genomic landscape. It is like fitting puzzle pieces together to reveal the bigger picture.
Example: Aligning the genomic data of an individual with Asian ancestry to a reference genome based on individuals of European descent might unveil unique genetic variations specific to the Asian population.
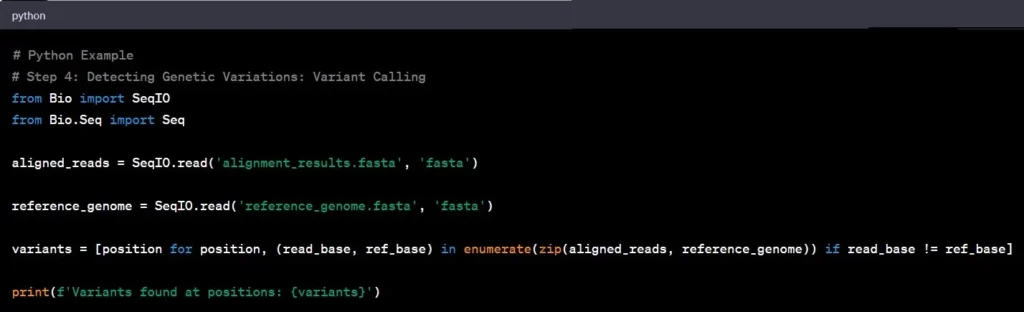
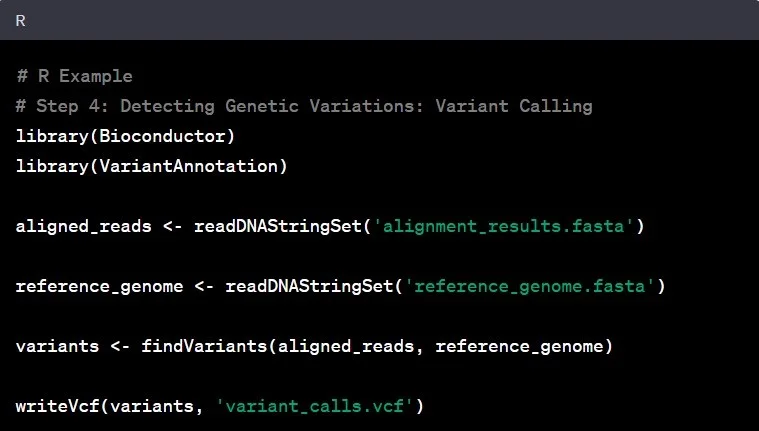
Step 4: Variant Calling
Variant calling involves identifying genetic variations – the differences between the aligned reads and the reference genome. Such as spotting single nucleotide polymorphisms (SNPs) and insertions/deletions (indels), by comparing aligned reads to the reference genome are common examples in genomic data analysis
Example: Identifying a single nucleotide polymorphism (SNP) in the BRCA1 gene can provide crucial information regarding breast cancer susceptibility.
Know more about R Bioinformatics Cheat Sheet for Beginners here.
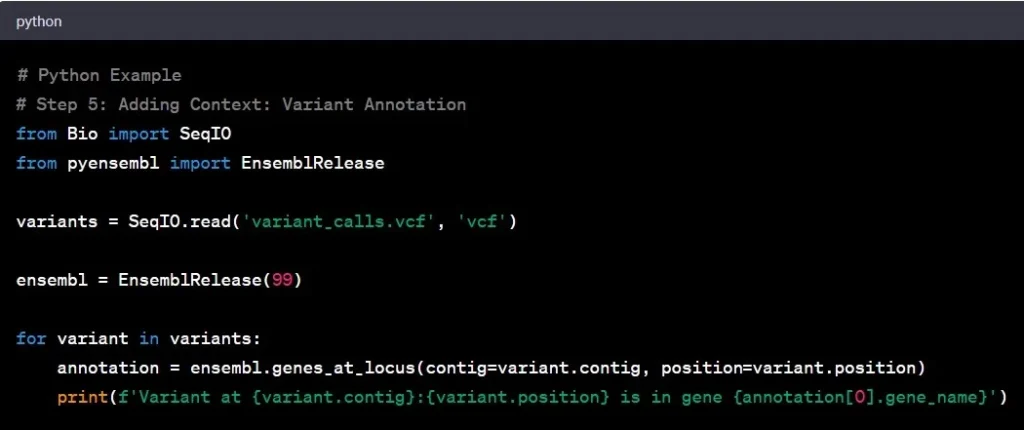
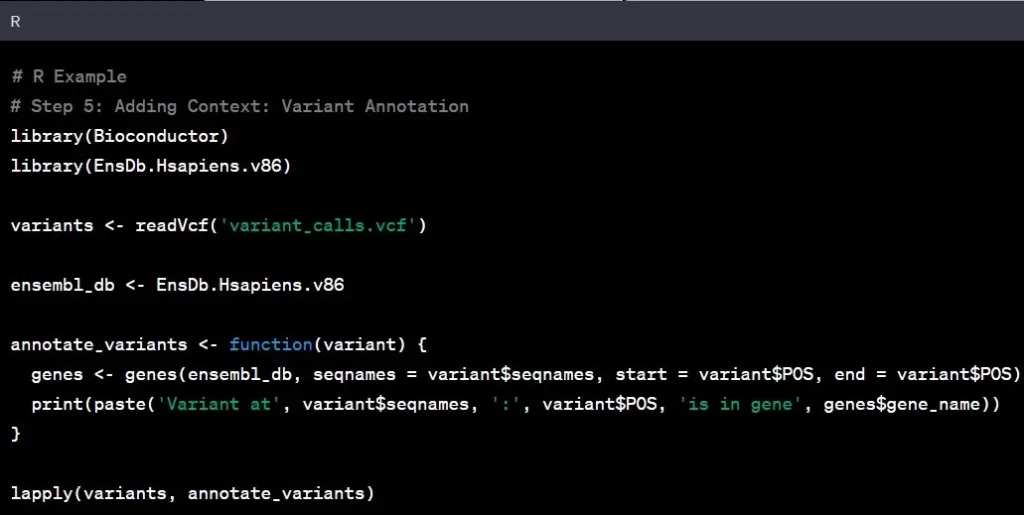
Step 5: Variant Annotation
Think of variant annotation as adding footnotes to the genomic text. It involves understanding the functional significance of identified variants and their potential impact on genes. It’s like deciphering the meaning behind the words in a book.
Example: Discovering a variant in a tumor suppressor gene may indicate a higher cancer risk, providing valuable insights for potential therapeutic strategies.
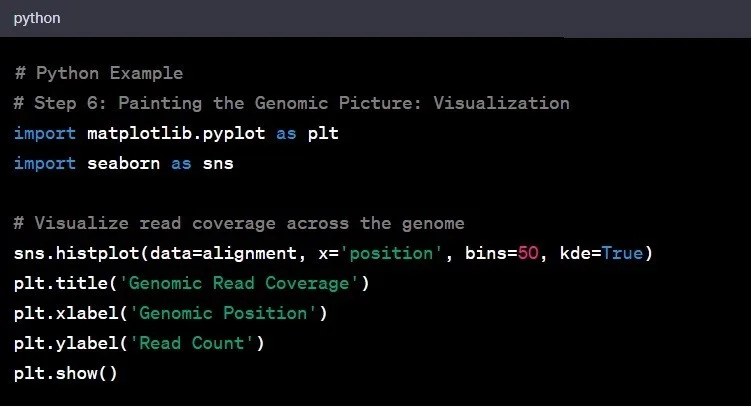
Step 6: Data Visualization
Now that we have our genomic data and insights, it’s time to visualize the findings. Visualization is the art of transforming raw data into meaningful stories. It is necessary step for genomic data analysis to gain insights and communicate findings effectively.
Example: Plotting the coverage across the genome could reveal regions with higher or lower sequencing depth, guiding further exploration.
Conclusion:
Genomic data analysis is a multifaceted process that involves acquiring, preprocessing, aligning, calling variants, annotating, and visualizing data. The seamless integration of Python and R in this guide provides a versatile toolkit for researchers and data scientists in the field. Mastering these steps and techniques not only enhance understanding of genomics but also contribute to advancements in the ever-evolving field of biological data science genomic research.
Click here to explore 11 Python Packages for Biological Data and Cheat Sheet.