Understanding gene expression is crucial in molecular biology. Two powerful tools used for this purpose are RNA sequencing (RNA-Seq) and microarrays. Both methods provide insights into how genes are turned on or off, but they do so in different ways. This article will compare and contrast RNA-Seq vs microarray, highlighting their strengths, limitations, and applications.
Basics of Gene Expression Analysis
What is Gene Expression?
Gene expression is the process by which information from a gene is used to create a functional product like a protein. Understanding which genes are active in different cells and conditions helps scientists understand biological processes and disease mechanisms.
Importance of Gene Expression Analysis
Analyzing gene expression allows researchers to identify which genes are involved in specific diseases, understand how genes interact, and develop new treatments. It’s a key area in research fields like cancer, immunology, and genetics.
What is RNA-Seq?
Definition and Process
RNA-Seq, short for RNA sequencing, is a technique that uses next-generation sequencing (NGS) to capture the transcriptome of a cell. It involves converting RNA to complementary DNA (cDNA), sequencing it, and then mapping the sequences back to a reference genome.
Advantages of RNA-Seq
- High Resolution: RNA-Seq provides a high-resolution view of the transcriptome, detecting even low-abundance transcripts.
- Broad Range: It can identify and quantify all RNA species, including mRNA, non-coding RNA, and small RNAs.
- Discovery Power: RNA-Seq can discover novel transcripts, splice variants, and gene fusions.
Limitations of RNA-Seq
- Cost: RNA-Seq can be expensive due to the need for high-throughput sequencing platforms and bioinformatics analysis.
- Complex Data Analysis: The data generated is complex and requires significant computational resources and expertise to analyze.
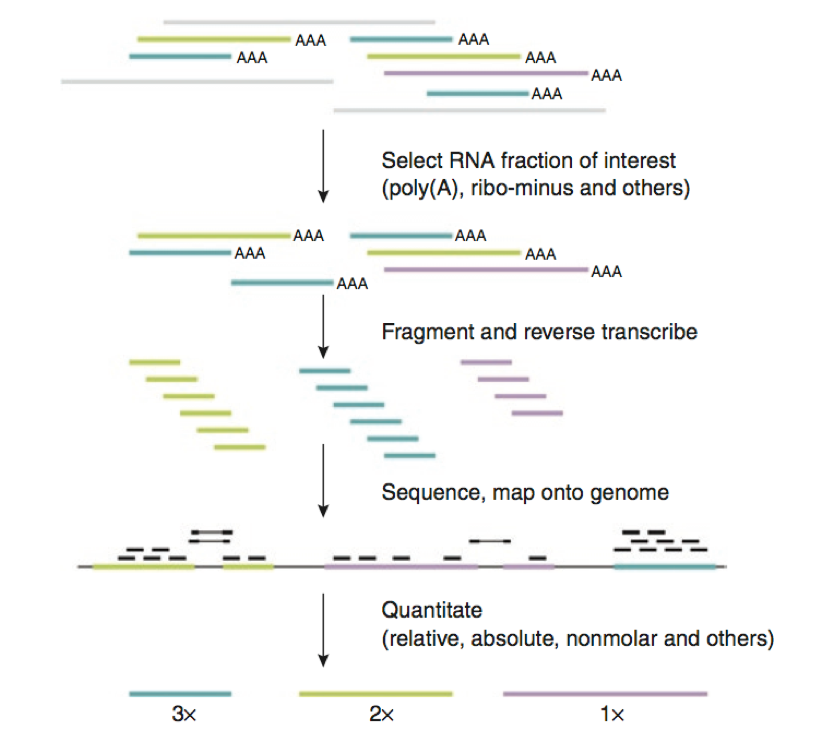
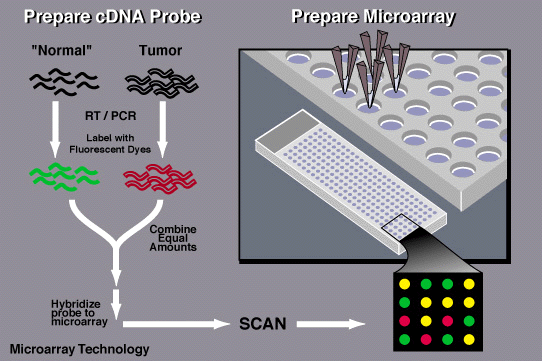
What is a Microarray?
Definition and Process
Microarrays are laboratory tools used to study many genes at once. They consist of a solid surface, like a glass slide, onto which DNA sequences are fixed in a grid. When RNA from a sample is labeled and applied to the microarray, it hybridizes with complementary DNA sequences, producing a signal that can be measured.
Advantages of Microarrays
- Cost-Effective: Microarrays are generally less expensive than RNA-Seq.
- Established Technology: They have been used for many years, and the technology is well-understood and widely available.
- Ease of Use: Microarrays are relatively straightforward to use and analyze.
Limitations of Microarrays
- Limited Dynamic Range: Microarrays have a limited dynamic range compared to RNA-Seq.
- Pre-Defined Probes: They rely on pre-defined probes, meaning they can only detect transcripts that are already known.
- Lower Sensitivity: Microarrays are less sensitive to low-abundance transcripts.
RNA-Seq vs Microarray: A Detailed Comparison
Sensitivity and Specificity
In the RNA-Seq vs microarray debate, RNA-Seq often wins in terms of sensitivity and specificity. RNA-Seq can detect a broader range of expression levels and is better at identifying low-abundance transcripts. Microarrays, while effective for many applications, may miss some of these lower-abundance signals.
Range of Detection
RNA-Seq has a wider dynamic range of detection. This means it can accurately measure both very high and very low levels of gene expression. Microarrays, on the other hand, have a more limited range and may not capture extreme expression levels as accurately.
Discovery of Novel Transcripts
One of the major advantages of RNA-Seq vs microarray is RNA-Seq’s ability to discover novel transcripts and splice variants. Because RNA-Seq does not rely on pre-defined probes, it can identify new and unexpected RNA sequences, providing deeper insights into the transcriptome.
Quantitative Accuracy
RNA-Seq provides more quantitative data compared to microarrays. The counts of RNA-Seq reads directly correlate with the abundance of the RNA molecules, making it a more precise method for quantifying gene expression levels.
Cost and Accessibility
While RNA-Seq offers many advantages, it is generally more expensive than microarrays. The high cost of sequencing and data analysis can be a barrier for some researchers. Microarrays, being less expensive and requiring less complex data analysis, remain a popular choice for many applications.
Data Analysis and Interpretation
The data generated by RNA-Seq is more complex and requires advanced bioinformatics tools for analysis. This can be both a strength and a limitation. For researchers with the necessary expertise and resources, RNA-Seq offers a wealth of information. However, for those without these resources, microarrays provide a simpler and more straightforward option.
Applications of RNA-Seq vs Microarray
Disease Research
In disease research, both RNA-Seq and microarrays are used to understand gene expression changes associated with diseases. RNA-Seq’s ability to detect novel transcripts and provide quantitative data makes it particularly valuable for identifying disease biomarkers and understanding disease mechanisms.
Drug Development
In drug development, understanding gene expression changes in response to treatments is crucial. RNA-Seq’s high resolution and quantitative accuracy provide detailed insights into how drugs affect gene expression, helping to identify potential therapeutic targets and understand drug mechanisms of action.
Personalized Medicine
Personalized medicine aims to tailor treatments to individual patients based on their genetic makeup. RNA-Seq’s ability to provide detailed and comprehensive gene expression profiles makes it a powerful tool for personalized medicine, helping to identify the best treatments for individual patients.
Functional Genomics
Functional genomics studies the functions and interactions of genes and proteins. Both RNA-Seq and microarrays are used in functional genomics to understand how genes are regulated and how they interact with each other. RNA-Seq’s ability to detect novel transcripts and provide quantitative data makes it particularly valuable in this field.
RNA-Seq vs Microarray: Practical Considerations
Experimental Design
When choosing between RNA-Seq vs microarray, researchers must consider their experimental design. RNA-Seq is ideal for discovery-based projects where the goal is to identify new transcripts or understand complex gene expression patterns. Microarrays, with their pre-defined probes and simpler analysis, are better suited for studies focusing on known genes and straightforward expression comparisons.
Sample Size and Replication
Sample size and replication are important factors in any gene expression study. RNA-Seq, while more expensive, can provide more detailed data from fewer samples. Microarrays, being less costly, allow for larger sample sizes and more replicates, which can improve the statistical power of a study.
Data Storage and Management
The data generated by RNA-Seq is large and requires significant storage and computational resources. Researchers must be prepared to handle and analyze large datasets. Microarrays generate smaller datasets, which are easier to store and manage, making them more accessible for researchers with limited resources.
Quality Control
Quality control is crucial in both RNA-Seq and microarray experiments. RNA-Seq requires careful handling of samples and rigorous quality control measures to ensure accurate results. Microarrays, while simpler, also require careful preparation and quality control to avoid issues like non-specific binding and background noise.
Future Directions
Advances in RNA-Seq
RNA-Seq technology is continuously evolving. Advances in sequencing technologies, such as single-cell RNA-Seq and long-read sequencing, are providing even deeper insights into the transcriptome. These advancements are likely to further increase the advantages of RNA-Seq vs microarray in the future.
Integration of Data
Integrating data from RNA-Seq and microarray experiments can provide a more comprehensive understanding of gene expression. Combining the strengths of both methods can help validate findings and provide a more complete picture of the transcriptome.
Cost Reduction
As RNA-Seq technology advances, costs are likely to decrease. This will make RNA-Seq more accessible to researchers and increase its use in various applications. Efforts to reduce the cost of sequencing and improve the efficiency of data analysis will play a key role in this trend.
Improved Bioinformatics Tools
The development of improved bioinformatics tools will make it easier to analyze and interpret RNA-Seq data. Advances in machine learning and artificial intelligence are likely to play a significant role in this area, helping researchers to make sense of complex datasets and uncover new insights into gene expression.
Conclusion
In the comparison of RNA-Seq vs microarray, each method has its own strengths and limitations. RNA-Seq offers higher sensitivity, a broader range of detection, and the ability to discover novel transcripts, making it a powerful tool for gene expression analysis. Microarrays, while less sensitive and reliant on pre-defined probes, are more cost-effective, simpler to use, and widely available.
The choice between RNA-Seq vs microarray depends on the specific needs of the research project, including the goals of the study, the available resources, and the level of detail required. By understanding the differences between these two methods, researchers can make informed decisions and choose the best tool for their studies.
As technology continues to advance, the landscape of gene expression analysis will continue to evolve. Both RNA-Seq and microarrays will play important roles in this evolution, contributing to our understanding of gene expression and advancing the fields of biology and medicine.