RNA sequencing (RNA-Seq) is a powerful technology used to study the transcriptome. The transcriptome is the complete set of RNA transcripts produced by the genome. It helps researchers understand gene expression patterns, identify new transcripts, and explore changes in gene expression under different conditions. Analyzing RNA-Seq data involves multiple steps, including data preprocessing, alignment, quantification, and differential expression analysis. One of the crucial steps in this process is visualizing the results of differential expression analysis, and a common tool for this is the volcano plot RNA-Seq.
What is a Volcano Plot?
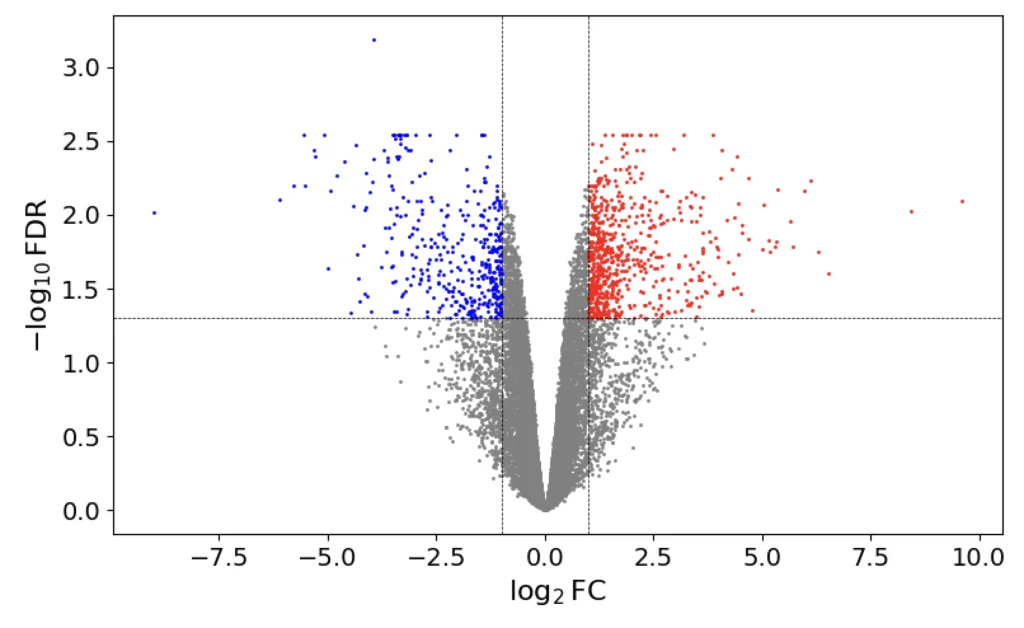
A volcano plot RNA-Seq is a type of scatter plot used to visualize the results of differential expression analysis. It helps identify genes that are significantly differentially expressed between different conditions. The plot gets its name from its shape, which resembles a volcano.
Structure of a Volcano Plot RNA-Seq
A volcano plot has two axes:
- The x-axis represents the log2 fold change (log2FC) of gene expression. This measures the difference in expression between two conditions. Positive values indicate upregulation, while negative values indicate downregulation.
- The y-axis represents the -log10 p-value. This measures the statistical significance of the observed changes in gene expression. Higher values indicate more significant changes.
Why Use a Volcano Plot RNA-Seq?
Volcano plot RNA-Seq is useful because it combines both statistical significance and magnitude of change. This allows researchers to quickly identify genes that are not only statistically significant but also have meaningful changes in expression. These plots make it easier to spot genes of interest for further investigation.
Generating a Volcano Plot from RNA-Seq Data
Creating a volcano plot from RNA-Seq data involves several steps. Let’s walk through the process step-by-step.
Step 1: Preprocessing the RNA-Seq Data
Before generating a volcano plot, the RNA-Seq data needs to be preprocessed. This includes quality control, trimming of low-quality reads, and alignment to a reference genome. Tools like FastQC, Trimmomatic, and STAR are commonly used in this step.
Step 2: Quantification of Gene Expression
After preprocessing, the next step is to quantify gene expression. This involves counting the number of reads that align to each gene. Tools like featureCounts or HTSeq can be used for this purpose. The output is typically a matrix of gene counts, with genes as rows and samples as columns.
Step 3: Differential Expression Analysis
The core step in creating a volcano plot is differential expression analysis. This involves comparing gene expression between different conditions to identify genes that are differentially expressed. Tools like DESeq2, edgeR, or limma are commonly used for this analysis.
Step 4: Creating the Volcano Plot
Once differential expression analysis is complete, the results can be visualized using a volcano plot RNA-Seq. Many software tools can generate volcano plots, including R (with the ggplot2 package), Python (with the matplotlib package), and dedicated bioinformatics tools like Galaxy.
Using R to Create a Volcano Plot
Here’s a simple example of how to create a volcano plot using R and ggplot2:
# Load necessary libraries
library(ggplot2)
# Assume 'results' is a data frame with columns 'log2FoldChange' and 'pvalue'
results <- read.csv("differential_expression_results.csv")
# Calculate -log10 p-value
results$negLogPval <- -log10(results$pvalue)
# Create the volcano plot
ggplot(results, aes(x=log2FoldChange, y=negLogPval)) +
geom_point() +
theme_minimal() +
labs(title="Volcano Plot RNA Seq",
x="Log2 Fold Change",
y="-Log10 P-value")
Step 5: Interpreting the Volcano Plot
Interpreting a volcano plot RNA-Seq involves identifying genes that are both statistically significant and have a meaningful fold change. Typically, genes with a high -log10 p-value and a high absolute log2 fold change are considered significant. These genes appear as points that are far from the origin, forming the “arms” of the volcano.
Applications of Volcano Plot RNA Seq
Volcano plots are widely used in RNA-Seq studies for various applications. Here are a few examples:
Identifying Biomarkers
In medical research, volcano plot RNA Seq can help identify biomarkers for diseases. By comparing gene expression in healthy and diseased tissues, researchers can pinpoint genes that are significantly upregulated or downregulated in the disease state. These genes can serve as potential biomarkers for diagnosis or treatment.
Studying Drug Effects
Volcano plots RNA-Seq are also useful in pharmacogenomics, where researchers study the effects of drugs on gene expression. By comparing gene expression before and after drug treatment, scientists can identify genes that respond to the drug. This information can help in understanding the drug’s mechanism of action and potential side effects.
Investigating Biological Pathways
Researchers often use volcano plot RNA-Seq to investigate biological pathways. By identifying differentially expressed genes, scientists can infer which pathways are activated or suppressed under certain conditions. This helps in understanding the biological processes involved in the condition being studied.
Challenges and Considerations
While volcano plots are powerful tools, there are some challenges and considerations to keep in mind.
Multiple Testing Correction
One of the main challenges in differential expression analysis is dealing with multiple testing. When testing thousands of genes, some may appear significant by chance. Techniques like the Benjamini-Hochberg procedure are used to adjust p-values and control the false discovery rate.
Choosing Cutoffs
Choosing appropriate cutoffs for fold change and p-value is crucial. Too stringent cutoffs may miss important genes, while too lenient cutoffs may include false positives. The choice of cutoffs depends on the specific research question and the context of the study.
Data Quality
The quality of RNA-Seq data can significantly impact the results. Poor quality data can lead to incorrect conclusions. It’s essential to perform rigorous quality control and preprocessing steps to ensure reliable results.
Advanced Visualization Techniques
While volcano plots are simple and effective, advanced visualization techniques can provide additional insights.
Interactive Volcano Plots
Interactive volcano plots allow researchers to explore the data more deeply. Tools like Plotly in Python or Shiny in R can create interactive plots where users can hover over points to see detailed information about each gene.
Integrating Additional Data
Integrating additional data, such as gene ontology or pathway information, can enhance the interpretation of volcano plots. Highlighting genes involved in specific pathways or functions can provide more context to the results.
Combining with Other Plots
Combining volcano plots with other types of plots can provide a more comprehensive view of the data. For example, heatmaps can show the expression patterns of significant genes across all samples, complementing the insights from the volcano plot.
Conclusion
Volcano plots are a valuable tool in RNA-Seq analysis. They provide a clear and intuitive way to visualize the results of differential expression analysis. By combining statistical significance and fold change, volcano plots help researchers identify key genes for further investigation.
Whether you are studying disease mechanisms, drug effects, or biological pathways, volcano plots can guide your analysis and interpretation. Despite some challenges, careful preprocessing, appropriate cutoffs, and advanced visualization techniques can enhance the utility of volcano plots in RNA-Seq studies.
As RNA-Seq technology continues to evolve, so will the tools and methods for analyzing and visualizing the data. Staying updated with the latest developments and best practices will ensure that you can make the most of volcano plots in your research.



